
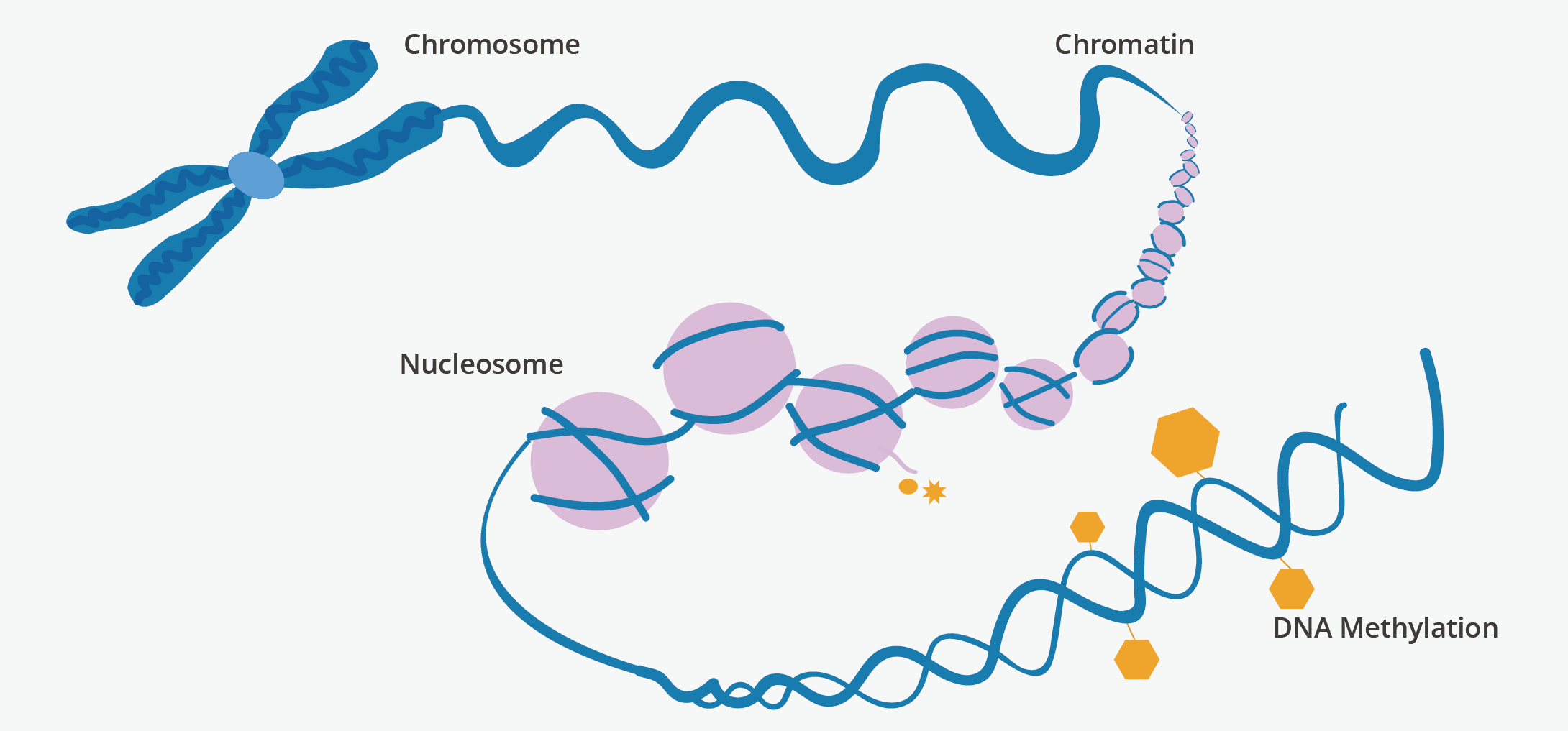
DNA methylation is a biological process that can change the activity of a DNA segment without altering its sequence. It is widely acknowledged as a heritable epigenetic factor that regulates gene activities. When located in a gene promoter, DNA methylation typically represses the transcription of the following gene.
Among the four DNA bases, cytosine and adenine can be methylated, resulting in modified form known as N6-Methyladenine (6mA), 5-Methylcytosine(5mC), and N4-Methylcytosine(4mC). Cytosine methylation is prevalent in both eukaryotes and prokaryotes, while Adenine methylation is relatively less common, even though it has been observed in bacterial, plant, and mammalian DNA.
DNA methylation plays an essential role in numerous important biological processes, including retroviral elements silencing, X-chromosome inactivation, genomic imprinting, suppression of repetitive element transcription and transposition, and more.
With the advancements in long-read sequencing technology offered by PacBio and Oxford Nanopore Technology (ONT), DNA methylation information now can be directly detected from the original DNA strands without the need for sodium bisulfite processing of DNA samples, which is a necessary step in whole genome bisulfite sequencing (WGBS) by the Illumina’s short-read sequencing technology.

Along with the high-quality long-reads sequenced by Novogene’s PacBio and ONT platforms, the methylation information will be delivered simultaneously. More details can be found in the table below.
Novogene’s Methylation Calling by PacBio and ONT Platforms
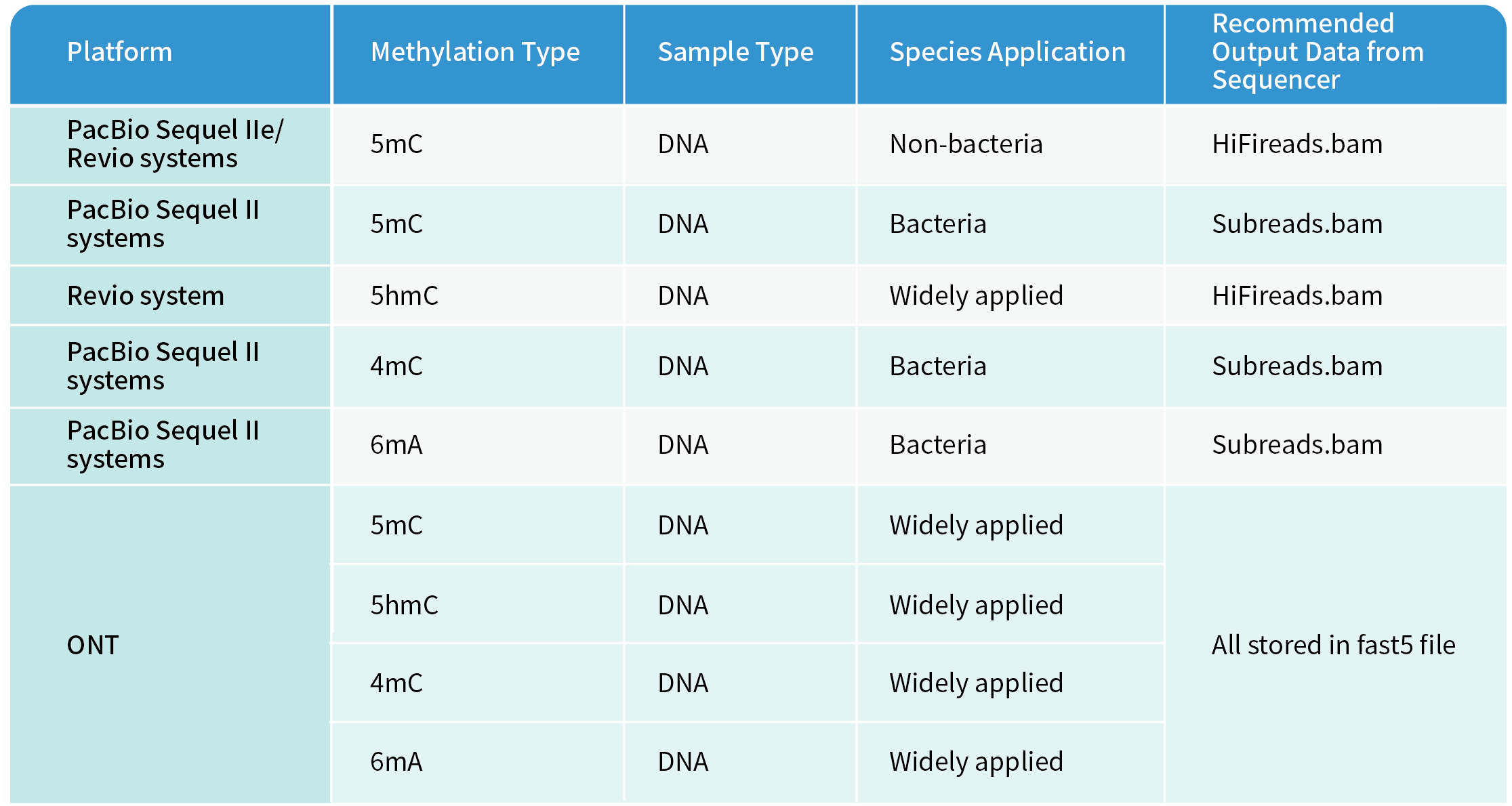
Notably, for the PacBio Revio system, since the Subreads.bam is not accessible to the sequencing machine, only HiFi reads containing 5mC methylation information will be delivered as the default setting. If other two kinds of methylation information, 4mC and 6mA, are required from the Revio system, the lab needs to make special adjustments to the sequencing system settings and export the original signal information, which will occupy larger storage. A reminder note for the special request is appreciated. Furthermore, the memory-consuming fast5 file from ONT will not be retained at Novogene for 30 days after data release as other data do. Instead, they will be kept at Novogene for a maximum of 21 days and automatically deleted within a week after downloading.

The Recommended Sequencing Depth for DNA Methylation Detection
The sequencing depth required for calling DNA methylation can vary between species, methylation locations in genome, and even tissue types. Below are the basic sequencing depths for reference, but adjustment may be necessary based on diverse research targets and purposes.
ONT: ≥10X for 5mC calling, ≥50X for 6mA and/or 4mC calling.
PacBio: ≥25X for 5mC calling, ≥100X for bacteria 6mA and/or 4mC.
Currently, Novogene can offer long-read sequencing data that also records the DNA methylation information sequenced using both PacBio and ONT platforms. However, we do not provide specific bioinformatics analysis services to further present DNA methylation types, contents and more. If such an analysis is needed, we are pleased to recommend several software options, including Primrose for PacBio data, and Nanopolish for ONT platform data, for your reference.
Let’s Talk about Your NGS Project
(Fields marked with an * are required)

Novogene Corporation Inc.
![]() 916-252-0068-383
916-252-0068-383
![]() inquiry_us@novogene.com
inquiry_us@novogene.com
![]() www.novogene.com
www.novogene.com
Copyright©2011-2023 Novogene Corporation
All Rights Reserved. Information and specifications are subject to change at any time without notice.
