
Summary
Brassica juncea is regarded as one of the earliest domesticated plants, which can be retrospected to as early as 3,000 BC mentioned as a condiment - mustard, in Sanskrit and Sumerian texts [1]. However, a clear understanding on evolutionary history of it has not been elaborated yet. Whether diverse ancient crop species was originated monophyletically or polyphyletically still remains controversial[7]. The relevant researches used to be hindered by limitations of sequencing technologies and complicated de novo assembly of repetitive, heterozygouse and polyploid angiosperm genomes[4]. While the advent of integrated sequencing strategies and hybrid assembly technologies, coupling with population genomics offers an opportunity to unveil its genetic dissection of the origin, domestication and diversification.
Here, we present a paper titled ‘‘Genomic insights into the origin, domestication and diversification of Brassica juncea’’ conducted by Lei Kang, Lunwen Qian, Ming Zheng, et al. from College of Agronomy, Hunan Agricultural University. This study integrated multi-omics approaches with technical assistance from Novogene. Leveraging the advancement of hybrid De novo sequencing, Resequencing and RNA sequencing, in alliance with population genomics, the authors not only reported a highly contiguous genome assembly version of super-scaffolds with an N50 value of 5.87 Mb[1], but also elucidated the complex evolutionary and domestication history, as well as human selection of candidate genes implicated in morphological diversification among diverse B. juncea subspecies[1].

Research Background

Brassica juncea(L.) Czern & Coss, a versatile and economically vital agricultural species, represents a newly formed allotetraploid (AABB, 2n=36) evolved from interspecific hybridization between two diploid progenitor species-B. rapa (AA, 2n=20) and B. nigra (BB, 2n=16)[2]. Cultivation of it, initiated since approximately 6,000 to 7,000 years ago and flourished from 2,300 BC onward[6], has been widely expanded to global spectrum of diverse environments as native plants, adapted crops and introduced weeds, spanning the continents of Asia, Europe, Africa, America and Australia[5].
Decoding the origin, domestication and morphological diversification of B. juncea and uncovering its genetic mechanisms on evolution of agriculturally conducive traits by polyploidy is crucial in identification of potential genetic driving forces contributed to selection response, which thereby will enable the prediction of the directional effects of selection in a polyploid crop genome[3]. Incorporated with superior chromosome-scale genome assembly, with improvement per genome size, contiguity and anchorage over the previous assembly version of stem[3] and Indian[2] mustard, this study is of major value to offer new perspectives for future research in breeding of this morphologically diverse condiment, oilseed, leaf, stem and root vegetable species[1].
Experiment Design and Research Pipeline
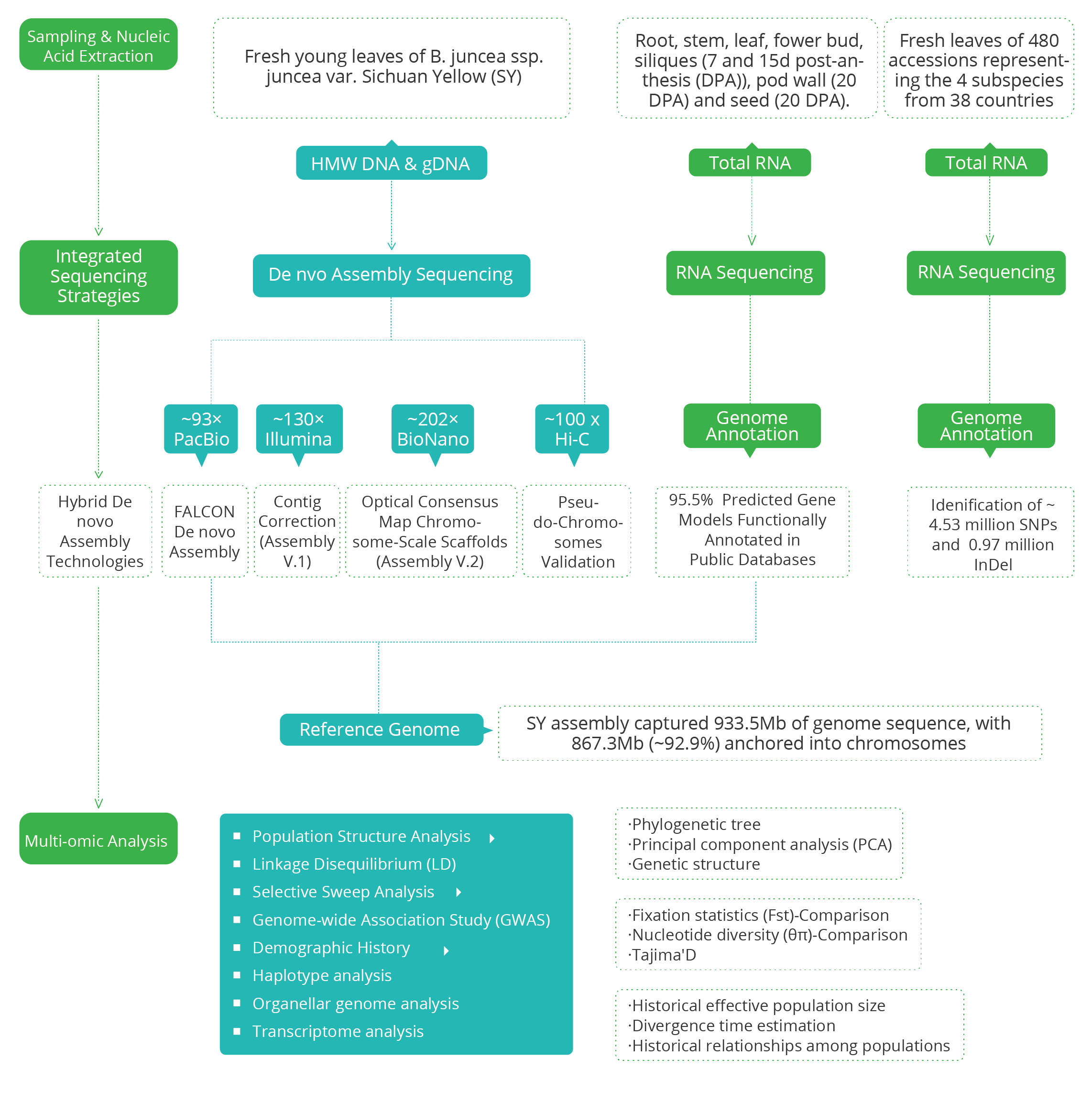
1.Integrated sequencing strategies and hybrid assembly technologies, involving PacBio long reads in conjunction with BioNano optical mapping and Hi-C chromatin interaction maps, were performed for a high-quality chromosome-scale genome assembly of the yellow-seeded B. juncea var. Sichuan Yellow (SY).
2.Re-sequencing was also conducted on both nuclear and organelle genomes (i.e. CP and mitochondrial (MT) genomes) of 480 accessions representing the 4 subspecies from 38 countries, which were self-pollinated over multiple generations prior to DNA extraction, to capture its full spectrum of genetic variation.
3.To further enable a detailed reconstruction of the evolutionary and domestication history of this ancient allotetraploid species, population genomics (i.e. Population Structure, Selective sweeps, GWAS analysis and Transcriptome analysis etc.) were applied, together with archaeological evidence and historical written records.
Discussion
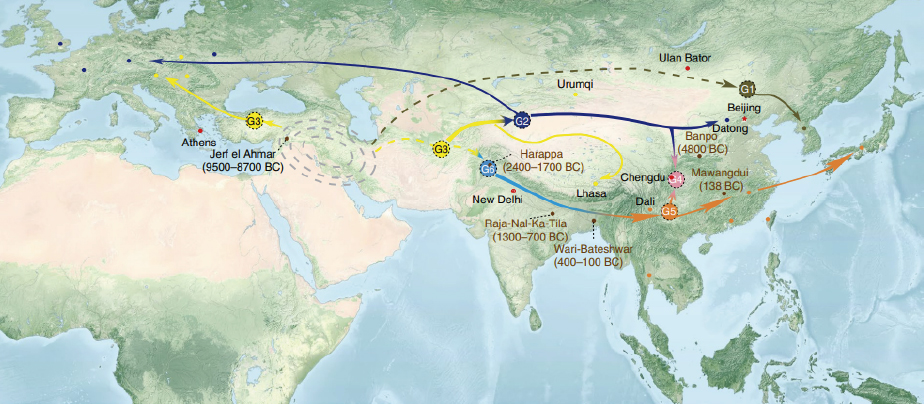
Nuclear and organelle phylogenies of 480 accessions worldwide advocated its monophyletic origin as early as 8,000–14,000 years ago in West Asia, via natural interspecific hybridization[1].
New crop types were evolved through spontaneous gene mutations triggered predominantly by natural hybridization and introgressions of at least three distinct clades along their independent domestication events of eastward expansion[1].
‘‘Seed mustard near Central Asia, oilseed mustard in the Indian subcontinent and root mustard in East Asia. As B. juncea spread eastward, yellow-seeded (Oriental) mustard arose in Northwest China, stem mustard in the Sichuan Basin and probably broad-leaf mustard in eastern India, by selection acting on via spontaneous mutations. Hybridization of leaf mustard with yellow-seeded and root mustard gave rise to early-maturing yellow-seeded mustard in the Yunnan–Kweichow Plateau and lobed-leaf mustard (var. mul-tisection Bailey) in eastern China, respectively.’’
Underlying genes and causal alleles for morphological variants are also identified, as potential contributors to root expansion, stem swelling, flowering time and seed size variation implicated in domestication and diversification[1].
Conclusion
Overall, integrating hybrid sequencing strategies with comprehensive bioinformatic analysis of Agrigenomics, this paper not only shed light on the biological issues of interests that used to be challenges for scientific researchers, but also offered constructive guidelines for genomics-based breeding and future relevant researches, thereby received extensive attention in the field of biological researches.
References
[1] Kang L. et al. Genomic insights into the origin, domestication and diversification of Brassica juncea. Nat Genet 53: 1392–1402 (2021).
[2] Paritosh, K. et al. A chromosome-scale assembly of allotetraploid Brassica juncea (AABB) elucidates comparative architecture of the A and B genomes. Plant Biotechnol. J. 19, 602–614 (2021).
[3] Yang, J. et al. Te genome sequence of allopolyploid Brassica juncea and analysis of diferential homoeolog gene expression infuencing selection. Nat. Genet. 48, 1225–1232 (2016).
[4] Michael, T.P. & VanBuren, R. Progress, challenges and the future of crop genomes. Curr. Opin. Plant Biol. 24, 71–81 (2015).
[5] Spect, C. E. & Diederichsen, A. Brassica in Mansfeld’s Encyclopedia of Agricultural and Horticultural Crops (ed. Hanelt, P.) 3, 1453–1456 (Springer Press, 2001).
[6] Prakash, S. & Hinata, K. Taxonomy, cytogenetics and origin of crop Brassicas, a review. Opera Bot. 55, 1–57 (1980)
[7] Vavilov, N. I. Phytogeographic basis of plant breeding. Chronica Bot. 13, 14–56 (1951).
Let's Talk about Your NGS Project
(Fields marked with an * are required)

Novogene Corporation Inc.
![]() 916-252-0068-383
916-252-0068-383
![]() inquiry_us@novogene.com
inquiry_us@novogene.com
![]() www.novogene.com
www.novogene.com
Copyright©2011-2022 Novogene Corporation
All Rights Reserved. Information and specifications are subject to change at any time without notice.
