ATAC-seq case study of discovering mechanism of the FOXA1 cistrome in treatment-emergent neuroendocrine prostate cancer
Application of multi-omics in cancer research

Prostate cancer is the second most common cancer in men worldwide. The development and progression of prostate cancer are driven by complex interactions between genetic and epigenetic factors. Epigenetic modifications, such as DNA methylation and histone modifications, regulate gene expression and chromatin accessibility without altering the DNA sequence. Chromatin accessibility reflects the dynamic state of chromatin structure and is associated with gene regulation and cellular identity.
ATAC-seq (Assay for Transposase-Accessible Chromatin using sequencing) is a cutting-edge technique that measures genome-wide chromatin accessibility by using a hyperactive Tn5 transposase to insert sequencing adapters into accessible regions of chromatin. ATAC-seq can reveal the locations of regulatory elements, such as promoters, enhancers, and insulators, and their interactions with transcription factors and other proteins. ATAC-seq can also identify cell-type-specific and disease-associated changes in chromatin accessibility and gene expression.

In this case study, Researchers applied ATAC-seq to investigate the epigenomic basis of lineage plasticity in prostate cancer, which is the ability of a cell to alter its identity and escape targeted therapy. Researchers focused on the role of FOXA1, a transcription factor that regulates androgen receptor (AR) signaling in prostate adenocarcinoma (PRAD), and how it is reprogrammed to neuroendocrine (NE) specific regulatory elements in neuroendocrine prostate cancer (NEPC). NEPC is an aggressive variant of prostate cancer that aberrantly expresses genes characteristic of NE tissues.
Researchers performed ATAC-seq on patient-derived xenografts (PDXs) of PRAD and NEPC, as well as on PRAD cells that were ectopically expressing the NE lineage transcription factors ASCL1 and NKX2-1. Researchers used bioinformatics tools to process the raw sequencing data, perform quality control, align reads to the reference genome, identify peaks of accessible chromatin regions, annotate peaks to genomic features, compare peak profiles between samples, perform differential accessibility analysis, identify transcription factor binding motifs, and perform gene ontology analysis.
Samples:
PRAD and NEPC cells from xenografts (PDXs); PRAD cells from cell lines
Library Preparation:
ChIP-seq, ATAC-seq, RNA-seq library
Sequencing Strategy:
Illumina platforms, paired-end 150 bp; Illumina Nextseq 500, paired-end 35bp
Bioinformatics Analysis:
Peak calling, enriched peaks analysis, Motif Enrichment analysis, Heatmap etc.
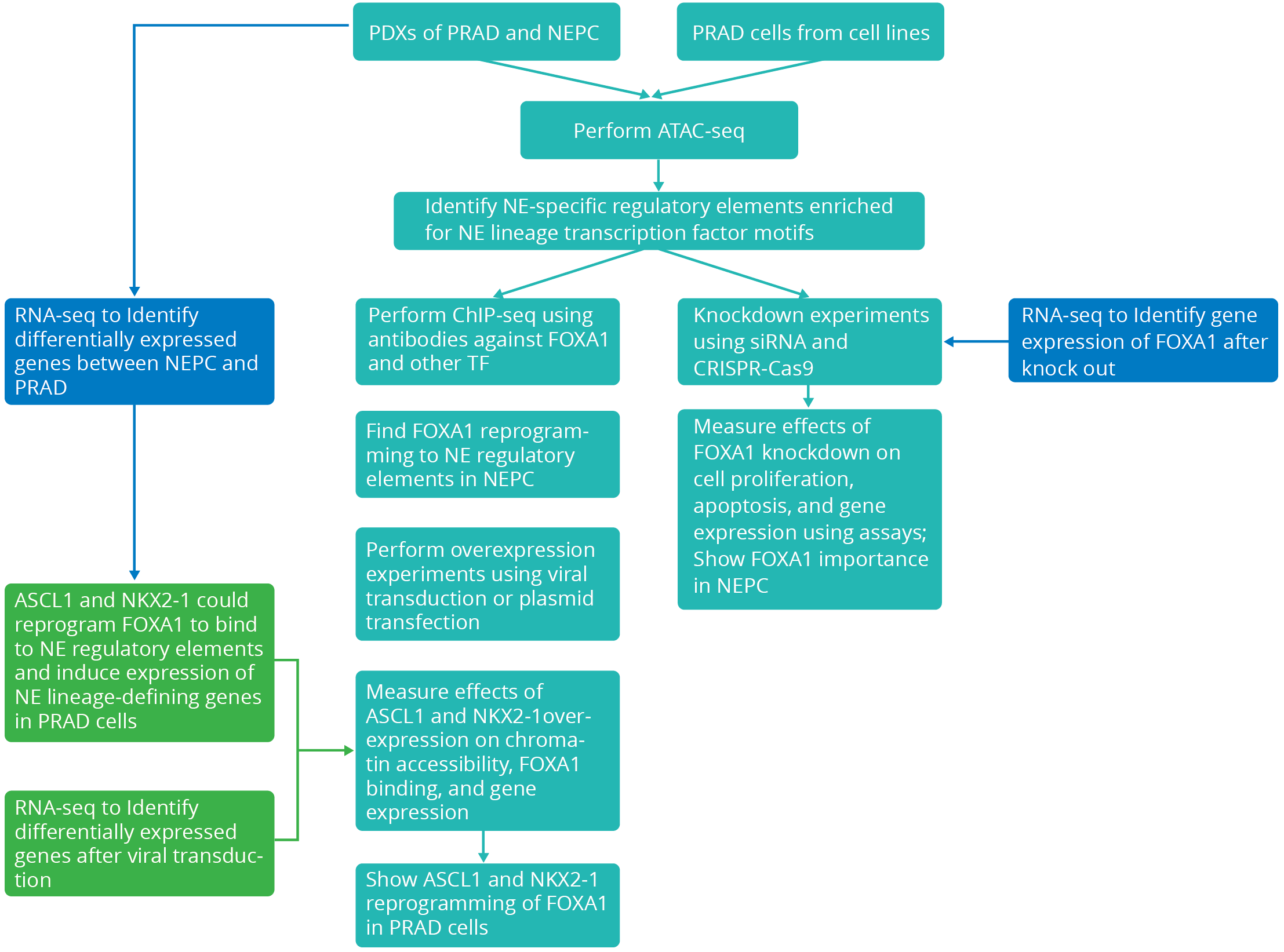
Major Research Strategy

Researchers identified a large network of cis-regulatory elements (~15,000) that were recurrently activated in NEPC. They discovered that FOXA1, a transcription factor that pioneers androgen receptor (AR) chromatin binding in PRAD, was reprogrammed to bind to these NE-specific elements in NEPC, independent of AR. FOXA1 was essential for NEPC proliferation and expression of NE lineage-defining genes. Moreover, researchers showed that ASCL1 and NKX2-1, two transcription factors that are characteristic of NE tissues, could reprogram FOXA1 to bind to NE elements and activate enhancer activity in PRAD cells. These findings reveal the epigenomic basis of lineage plasticity and therapeutic resistance in prostate cancer.

This reference illustrates the importance of FOXA1 in NEPC and provides a principled approach to identifying cancer dependencies through epigenomic profiling. It also showcases the potential of ATAC-seq as a powerful tool for studying the epigenomic landscape of prostate cancer. ATAC-seq can reveal regions of chromatin that are differentially accessible between sample subtypes, which may reflect changes in transcription factor binding and gene expression. Another strength of this reference is the combined application of multiple epigenetic and transcriptomic sequencing methods. By using ATAC-seq as a primary tool to locate differentially accessible regions, the researchers then use ChIP-seq and RNA-seq to validate the binding and expression of FOXA1 and its target genes. They also use knock-out experiments to confirm the functional role of FOXA1 in NEPC. This reference demonstrates how multi-omics can be integrated to reveal the molecular mechanisms underlying lineage plasticity and therapeutic resistance in prostate cancer.

Baca, S. C., Takeda, D. Y., Seo, J.-H., Hwang, J., Ku, S. Y., Arafeh, R., Arnoff, T., Agarwal, S., Bell, C., O’Connor, E., Qiu, X., Abou Alaiwi, S., Corona, R. I., Fonseca, M. A. S., Giambartolomei, C., Cejas, P., Lim, K., He, M., Sheahan, A., … Freedman, M. L. (2021). Reprogramming of the FOXA1 cistrome in treatment-emergent neuroendocrine prostate cancer. Nature Communications, 12(1), 1979. https://doi.org/10.1038/s41467-021-22139-7
Let’s Talk about Your NGS Project
(Fields marked with an * are required)

Novogene Corporation Inc.
![]() 916-252-0068-383
916-252-0068-383
![]() inquiry_us@novogene.com
inquiry_us@novogene.com
![]() www.novogene.com
www.novogene.com
Copyright©2011-2023 Novogene Corporation
All Rights Reserved. Information and specifications are subject to change at any time without notice.
